We are one of the largest cancer centres in the UK and provide some of the most up-to-date treatment, care and support for people with cancer.
Leeds Cancer Centre
Leeds Cancer Centre diagnoses and treats cancer for the people of Leeds and the Yorkshire region.
-

What the Leeds Cancer Centre do
Our services enable us to provide the most advanced treatment, care and support for those affected by cancer.
-

Cancers and their treatments
An A-Z list of the different types of cancers and their treatments.
-
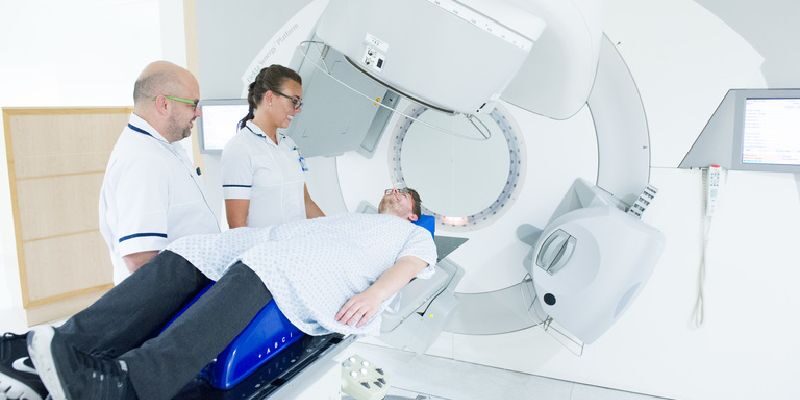
Diagnostic and treatment services
This section provides information on the range of diagnostic and treatment services available.
-
Leeds Cancer Support
Leeds Cancer Support offer information and a wide range of support in a warm and welcoming environment.
-
Urgent referral from a GP
Information if you have been referred by your GP to the Leeds Cancer Centre.
-
Health and wellbeing
Information on maintaining a sense of wellbeing with a cancer diagnosis.
-
The Thinking Ahead programme
Thinking Ahead is a Health and Wellbeing Programme for patients living with incurable cancer, who may or may not be receiving treatments, as well as their family members.
-
Patient experience
Information on how you can feedback your experience of cancer care.
-
Maggie’s Centre
An oasis of calm offering the best possible support free to anyone with cancer and their families.
-
Contact us
How to get in touch with the Leeds Cancer Centre.
-
Patient hotel and useful information
Find out more about the patient hotel and other available facilities.